Fungal Spore Ejection
Background
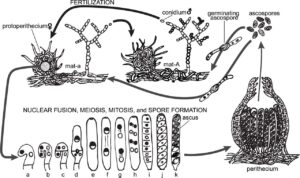 Understanding the mechanisms of discharge and dispersal of fungal spores are important for understanding the spread of disease caused by many fungal pathogens as well as evolutive mechanisms of survival. Adaptations in spore shape and launch mechanism [1,2] to exit the boundary layer for passive dispersal by the wind [3] have been studied in several species. One important factor is the role of surface proteins (hydrophobin) that make the spores hydrophobic [2,4-7] which mediates their interactions with the environment and may affect their dispersal success. In the model fungal species Neurospora crassa, the mutant easily wettable (eas), also known as circadian clock-controlled gene-2 (ccg-2), has been characterized intensively [8,9]. Genetic tools available to manipulate N. crassa make it a good candidate forcarefully studying the relationship between genetic traits and success in dispersal, as may lead to strategies for inhibiting the spread of fungal diseases.
Understanding the mechanisms of discharge and dispersal of fungal spores are important for understanding the spread of disease caused by many fungal pathogens as well as evolutive mechanisms of survival. Adaptations in spore shape and launch mechanism [1,2] to exit the boundary layer for passive dispersal by the wind [3] have been studied in several species. One important factor is the role of surface proteins (hydrophobin) that make the spores hydrophobic [2,4-7] which mediates their interactions with the environment and may affect their dispersal success. In the model fungal species Neurospora crassa, the mutant easily wettable (eas), also known as circadian clock-controlled gene-2 (ccg-2), has been characterized intensively [8,9]. Genetic tools available to manipulate N. crassa make it a good candidate forcarefully studying the relationship between genetic traits and success in dispersal, as may lead to strategies for inhibiting the spread of fungal diseases.
In this research we will study the mechanical constraints of this mechanism by detailed measurement of the launching process, and physical properties of different specimens.
IRES student involvement
Ascospore ejection will be recorded using different techniques of high-speed, high-magnification imaging with laser illumination in controlled flow conditions. Spores recaptured at varying distances from the perithecia will be analyzed via optical microscopy and SEM to determine the point of ascospore breakup, characterize spore shape, and unveil the physical mechanism involved.
This project aims to characterize the discharge of ascospores (cell packets of 8 spores) from the fruiting bodies (perithecia) of N. crassa and their eventual breakup and dispersal into free stream flow. Dr. (Kwangwon) Lee will advise a student on the mycological background of the project at Rutgers, where samples of wild type and eas N. crassa will be grown with natural and synthetic media prior to travel. In Chile, ascospore ejection will be recorded using high speed, high magnification imaging with laser illumination in controlled flow conditions. Spores recaptured at varying distances from the perithecia will be analyzed via visual microscopy and SEM to determine point of ascospore breakup and characterize spore shape, respectively.
References
[1] Roper, M., Pepper, R. E., Brenner, M. P. & Pringle, A. Explosively launched spores of ascomycete fungi have drag-minimizing shapes. Proc Natl Acad Sci, 2008.
[2] Fritz, J. A., Seminara, A., Roper, M., Pringle, A. & Brenner, M. P. A natural O-ring optimizes the dispersal of fungal spores. J R Soc Interface, 2013.
[3] Roper, M. et al. Dispersal of fungal spores on a cooperatively generated wind. Proc Natl Acad Sci, 2010.
[4] Luciano-Rosario, D. et al. The Hydrophobin Gene Family Confers a Fitness Trade-off between Spore Dispersal and Host Colonization in Penicillium expansum. mBio, 2022.
[5] Macindoe, I. et al. Self-assembly of functional, amphipathic amyloid monolayers by the fungal hydrophobin EAS. Proc Natl Acad Sci , 2012.
[6] Trail, F. Fungal cannons: explosive spore discharge in the Ascomycota. FEMS Microbiol Lett, 2007.
[7] Talbot, N. J. Aerial morphogenesis: enter the chaplins. Curr Biol, 2003.
[8] Bell-Pedersen, D., Lewis, Z. A., Loros, J. J. & Dunlap, J. C. The Neurospora circadian clock regulates a transcription factor that controls rhythmic expression of the output eas(ccg-2) gene. Mol Microbiol, 2001.
[9] Arpaia, G., Loros, J. J., Dunlap, J. C., Morelli, G. & Macino, G. The interplay of light and the circadian clock. Independent dual regulation of clock-controlled gene ccg-2(eas). Plant Physiol,1993.