Research
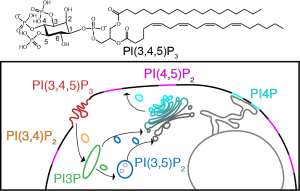
Phosphatidylinositol (PIPs)
Phosphatidylinositol phosphates (PIPs) are a class of mammalian lipid that are based off of phosphorylation of the headgroup of phosphoinoisitol (PI).
Mis-regulation of PIP pathways leads to disease (even cancer)
Bind with specificity to 100s of proteins, including PH domains
Goal: Establish a ssNMR methodology for observing PIPs and their interactions with Kindlin-2 (K2) PIP3-binding domains to gain insight into structural and dynamical changes which occur in the lipid and protein
The atomic resolution insight provided by solid-state NMR is vital for expanding our understanding of PIP-protein binding. In particular, we utilize 1H and 31P for studying our lipid systems. Both are naturally 100% abundant, therefore not needing any further labeling, and have high gyromagnetic ratios, a measure of magnetic susceptibility.
We seek to study membrane proteins in their native lipids, and determine the effects the lipids bilayer itself has on the protein and understand why they occur via the direct investigation of membranes by solid-state NMR. To do this, we continue to develop techniques for directly probing the structure and dynamics of lipid bilayers by solid-state NMR as well as the effects that different lipid species and small molecules can have on them. At Rutgers we have improved the 31P spectroscopy used to directly observe lipid head-groups. We use very-fast MAS and proton detection to observe lipid-lipid and lipid-protein interactions. We are developing pulse sequences and data analysis workflows to measure precise lipid-protein distances using 1H, 13C, 15N and 31P.
Figure: Top: A PIP, Bottom: The pathway used to probe structural and dynamic changes that occur in lipids and proteins.
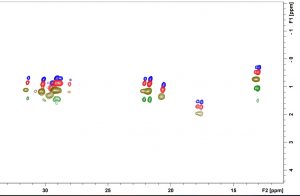
Structure and Dynamics of Ionic Liquids
Ionic Liquids are molten salts that are liquid at or near room temperature. The structure of IL leads to the existence of separate chemical environments in the bulk liquid. Parts of the liquid that exists within these environments exhibit distinct structural and dynamic properties. This leads to microscopic heterogeneity in the bulk structure of the liquid.
Through the use Solid State Magic Angle Spinning Nuclear Magnetic Resonance (ssMAS NMR) and high energy X-Ray Scattering experiments we can interrogate the sub-ion and bulk structural environments present in the liquid. Measurement of nuclei dependent relaxation times allow us to probe the individual dynamics of different parts of the IL. Combination of these techniques allows for the elucidation of the complete catalog of structural and dynamic features present in the liquid. This project is a collaboration with Prof. (emeritus) Edward W. Castner, from Rutgers University, Prof. Claudio Margulis’ group from the University of Iowa, Prof. Sharon Lall-Ramnarine group from Queensborough Community College, Prof. Hideaki Shirota from Chiba University Japan and Dr. James Wishart group from Brookhaven National Laboratory.
2D HC INEPT experiments reveal different carbon sites of IL lose mobility at different rates when cooling. This behavior is further proven by the non-uniform decrease in the T2 relaxation times of different carbon sites.
Figure: The 1H, and 13C, 2D NMR spectra of Phosphonium(14,666) and NTf2 at different temperatures.
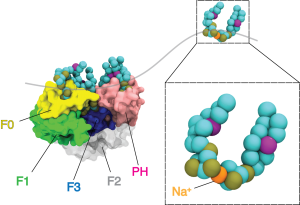
Molecular Dynamics to Understand Membrane Properties
In conjunction with our ssNMR studies of biomimetic membranes, we use multiresolution molecular dynamics (MD) simulations to understand the binding mechanisms of peripheral membrane proteins (PMPs) to lipid bilayers. This involves coarse grained through quantum-scale simulation of PMPs and their effector lipids. More specifically, we are interested in the Integrin signaling pathway which has been implicated in diverse disease processes including aggregation of inflammatory cell recruitment and tumor metastasis. Kindlin-2 is a protein involved in initial integrin signaling events and has been found to enhance talin-mediated activation of integrin. However, the basis for recruitment of Kindlin-2 by phosphatidylinositol phosphates (PIPs) and its impact on Integrin activation is still poorly understood. We employ computational methods to study the biochemical factors which effect PIP dynamics at the lipid bilayer and their role in PMP binding.
Figure: Course grained simulations of K2 bound to PIP containing membranes. Using the INSANE framework for course graining, we were able to run longer simulation times with CPUs nodes. This allows us to sample inter-domain orientations relative to the bilayer.
NMR Method Development
We seek to apply the power of 1H-detected ssNMR to growing numbers of areas in ssNMR. Due to the fact 1H’s gyromagnetic ratio is 4 times that of 13C, experiments detecting 1H are more sensitive and therefore faster. While 1H detection is increasing well developed in terms of chemical shift assignments, there is still a wide array of areas where it has not yet been deployed. In particular, we are interested on using ultrafast MAS with 1H detection to gain structural insight without deuterated samples. We are developing new pulse sequences to determine intermolecular distances both within a protein and intermolecularly, as well as study dynamics at many timescales. We have a MAS probe which can spin at 40 kHz and another which spins at 100 kHz is on the way.
In addition to 1H detected methods, we will use 31P detected methods. 31P is spin 1/2, 100% abundant, and relatively high gamma (17.235 MHz/T). It has good lineshape and sensetivity, a wide chemical shift range, and a large dipolar coupling range (~ 8 A). In addition, we expect to be able to filter out the lipid phosphate signal to obtain spectra with good signal-to-noise of specific phosphates we are interested in.