Sample Processing
Once environmental samples are collected, there are a couple of molecular pipelines that can be used to analyze samples.
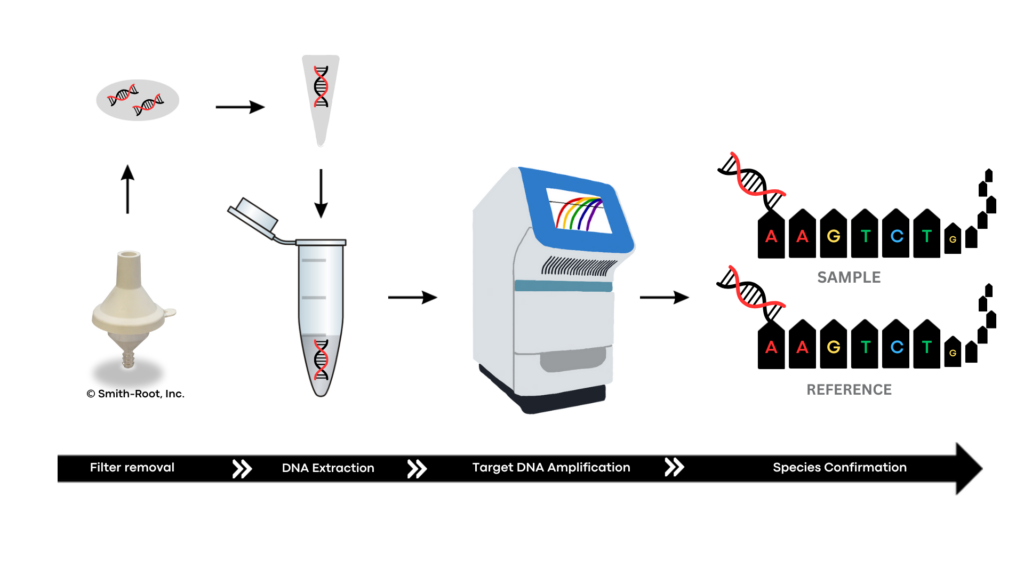
Filter Pulling and Extraction
To start any molecular workflow, the eDNA we’ve collected must be extracted from the filter. This process begins with pulling the filter from its housing, placing it into a tube, and using commercial reagents or kits or processes to extract the DNA sample from the filter. This extraction process allows us to isolate DNA from the filter and potentially remove compounds that may interfere with sample analysis.
At this point, DNA can enter one of two different pipelines:
Quantitative PCR (qPCR)
Analyzing our eDNA samples using qPCR technology allows us to determine the presence, or absence, of a single target species. qPCR assays are extremely sensitive molecular tests designed to amplify and detect sequences of DNA unique to a target species. By designing and utilizing a species-specific qPCR assay, we can infer the recent presence or absence of a particular species at a sampling location.
Metabarcoding
Similar to qPCR, the metabarcoding pipeline relies on DNA amplification, but this pipeline amplifies DNA sequences of interest in an eDNA sample for a group of target organisms, revealing a list of DNA sequences that can be matched to reference databases to identify the organisms present within each sample collected. This pipeline can be used to identify a majority of the insects, fish, or mammals present in an ecosystem.
.