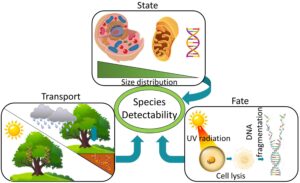
What is eDNA?
Environmental DNA allows for the collection of valuable biodiversity information across global ecosystems by taking advantage of one simple fact of life: all organisms shed their DNA, (via mucus, feces, exuviae, skin, hair, or reproductive cells, etc.), into the environment around them. This environmental DNA, or eDNA, can be easily captured and used to detect an organism’s recent presence in a location without the need for visual detection or capture. The likelihood of DNA detection depends on environmental conditions and the state of the DNA when it’s shed. UV radiation, microbial activity, high temperatures, heavy precipitation, low pH, and fast water currents can all influence the persistence and presence of DNA in the environment. DNA inside intact cells is less susceptible to degradation via these factors; free DNA may degrade more quickly. All of these factors affect how our lab collects and utilizes eDNA in a given environment.
How does our lab use eDNA ?
-
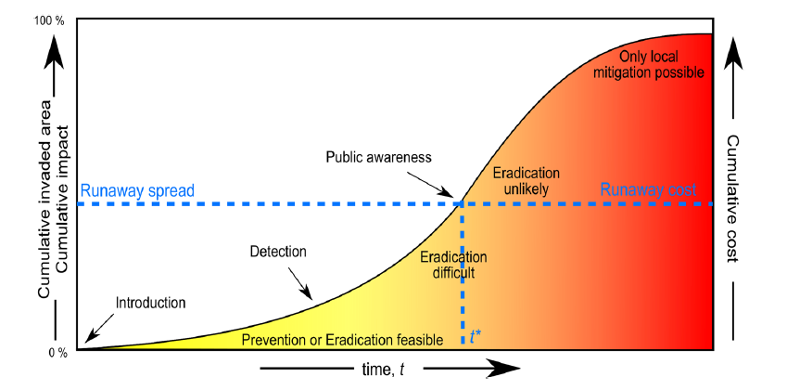
Ahmed et al., 2022 Non-native species introductions have risen dramatically in proportion with increasing global trade and travel (Seebens et al., 2017; Sikes et al. 2018). Control and eradication techniques for the subset of these species that cause ecological, economic, or cultural harm are consistently more effective when applied to small, localized populations (Ahmed et al., 2022). However, initial species introductions can often go unnoticed, because finding anything that is rare is difficult, costly, or time-intensive. eDNA surveys can be used to address this dilemma, allowing invasive species detection while populations are still at very low abundance. With proper monitoring, this rapid initial detection can give managers a head-start in containing and potentially eradicating an invasive species.
-

Anthropogenic changes to the environment have caused extinction rates to rise significantly above background rates (Pimm et al., 2014), demonstrating greater need for monitoring and conservation efforts. However, threatened, rare, or elusive species can be very difficult to detect via traditional sampling methods, often requiring major investments in on-the-ground surveys and physical capture of individuals that we seek to protect. Species-specific eDNA surveys can detect the presence of such species ‘sight unseen’, giving conservation practitioners the opportunity to protect vulnerable or endangered species.
-

Traditionally, biodiversity assessments have been conducted using techniques such as visual identification, physical capture, acoustic monitoring, or camera trapping. These methods can be costly or time-consuming and are often restricted for use to a subset of all species utilizing a site of interest. By collecting eDNA samples, we can use metabarcoding techniques to establish the recent presence of a wide range of species in a particular ecosystem or habitat.
Future of eDNA

Air eDNA
Recent publications have demonstrated species detection by simply filtering DNA suspended in the air. A study conducted at the Copenhagen Zoo successfully detected numerous exhibit species, feed species, pest species, and wild species (Lynggaard et. al., 2022). Some preliminary successes with airDNA have prompted experimentation using existing air quality monitoring infrastructure (e.g. air-quality networks) (Clare et. al., 2022; Littlefair et. al., 2023). We look forward to expanding the use of airDNA in our own work related to invasive species detection in forested and agricultural ecosystems.

Flower eDNA
Flowers are now being utilized by some groups as a substrate that regularly collects DNA from pollinators and other terrestrial arthropod communities (Thomsen & Sigsgaard, 2019; Newton et. al., 2023). Studies that aggregated DNA from flowers have had success in detecting a range of arthropod species (pollinators, predators, parasitoids), and determining patterns in plant-insect interaction webs.