Research
1. Next-generation CRISPR technologies: Developing Therapeutic Base Editing Agents for Genetic Diseases and Cancer.
Over 6,000 human genetic diseases are caused by a point mutation. Recent development in CRISPR technology generates hope for a cure for those diseases. Current CRISPR- based gene-editing approaches rely on generating double-strand DNA breaks (DSBs). DSBs can either induce non-homologous end joining (NHEJ) to generate a frameshift mutation to inactivate the target gene or trigger homology-dependent repair (HDR) to correct a mutation. Unfortunately, these approaches have two main limitations: somatic cells do not have HDR activity or have very low HDR activity, and DNA double-strand breaks are highly oncogenic.
Base editing is a precision genome technology that can introduce targeted nucleotide modifications without the need to generate DSB at the target site. This is achieved by the recruitment of a DNA deaminase, such as a cytidine deaminase, to a nuclease-deficient Cas9/sgRNA complex. Pin-point™ (Fig. 1) is a modular base editing system in which the effector and Cas9 are separate molecules. The effector module is recruited to the CRISPR/sgRNA complex by a chimeric RNA molecule containing an RNA aptamer (Fig. 2). Pin-point, invented in 2015, can be used to correct a loss of function mutation or install premature stop codons for therapeutic gene targeting (knockout) (Fig. 3) (Collantes et al., 2021).
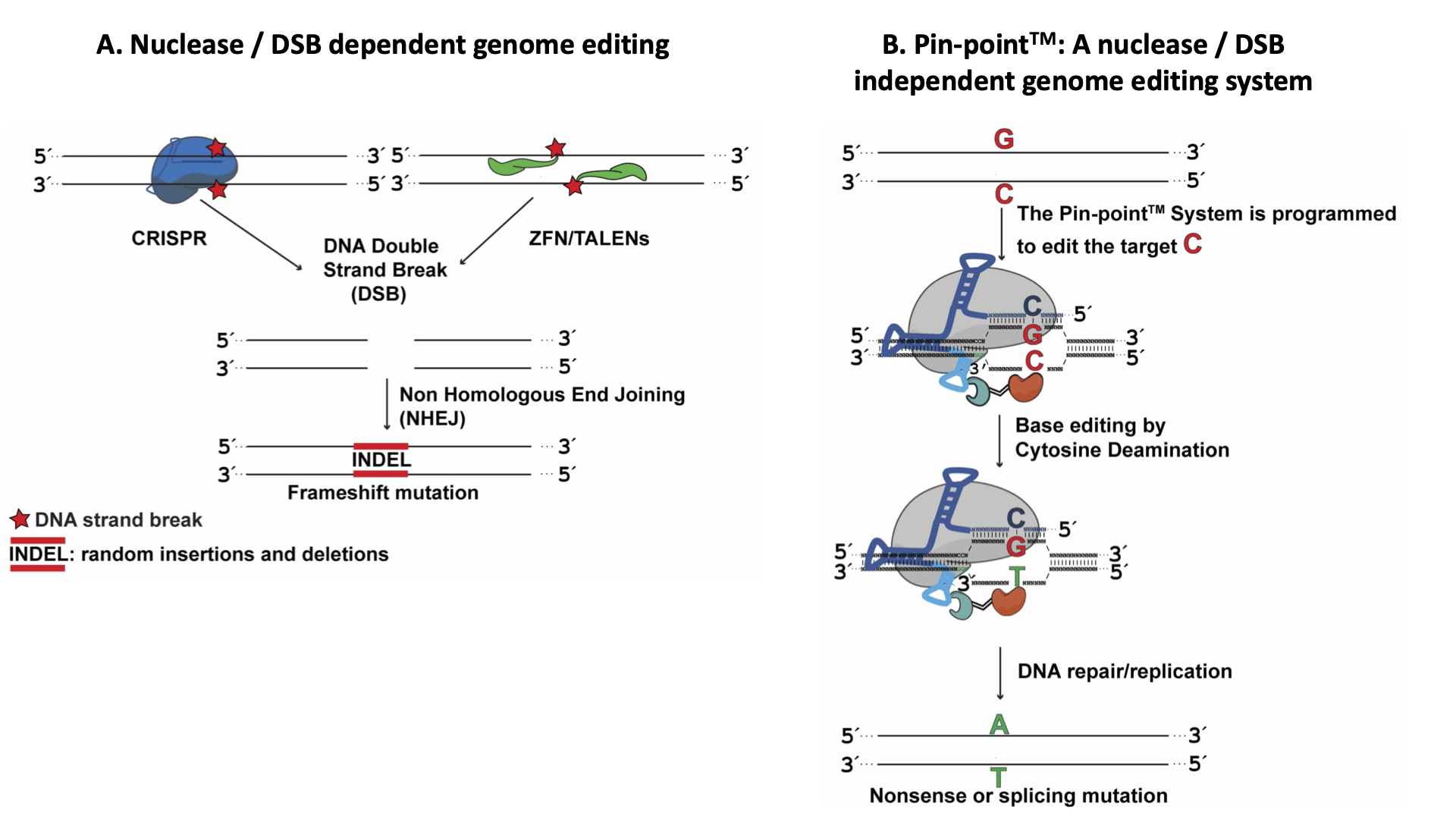
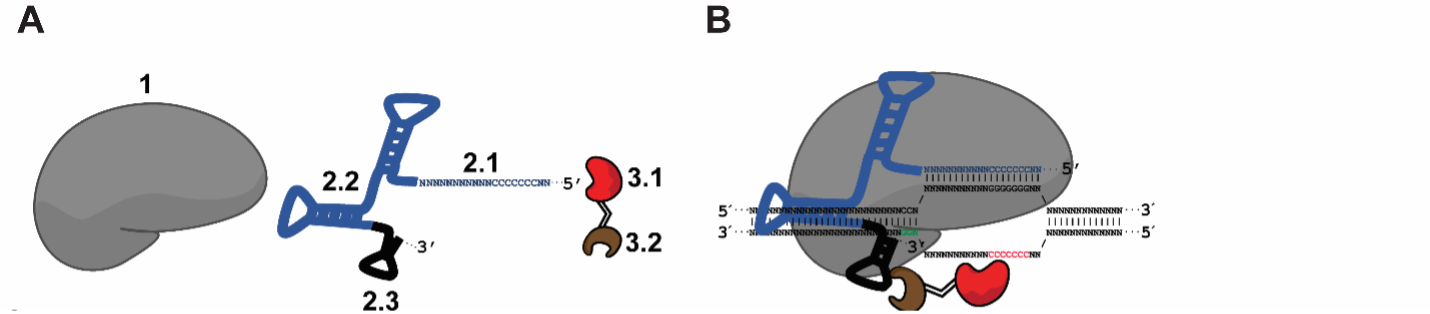
Figure 2. (A) Components of the Pin-point platform, from left to right: 1 Sequence targeting component dCas9 or Cas9 nickases, 2 Chimeric RNA scaffold containing a guide RNA motif (for sequence targeting; 2.1), CRISPR motif (for Cas9 binding; 2.2), and recruiting RNA aptamer motif (for recruiting the effector-RNA binding protein fusion; 2.3), and 3 Fusion protein consisting of the effector cytidine deaminase (3.1) fused to an RNA aptamer ligand (3.2) (B) Schematic of the modular complex at the target sequence (Collantes et al., 2021).
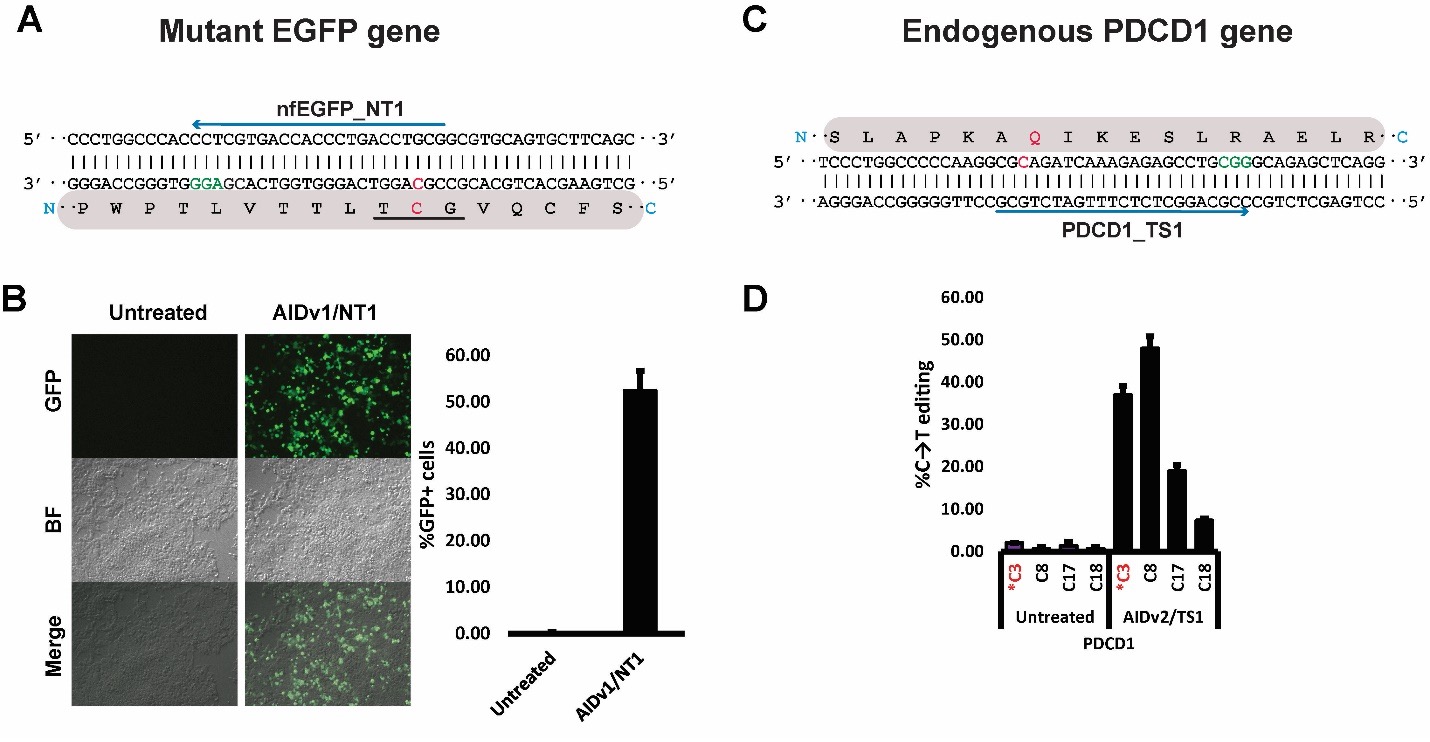
Figure 3 Applications of Pin-point Base Editing system in human cells. (A) Mutant non-fluorescent EGFP (nfEGFP) target region. A→G loss-of-function mutation at the chromophore sequence renders the EGFP protein non-fluorescent. The mutant chromophore is underlined in black. One gRNA targeting the non-template strand (nfEGFP_NT1) is shown as a blue arrow, the PAM sequence is shown in green, and the target cytosine is shown in red. The corresponding protein sequence is shaded in gray. (B) Treating the nfEGFP gene with Pin-point efficiently restores the fluorescence. Panels show representative sections of plates under a fluorescence microscope after the indicated treatments. The bar graph shows flow cytometry analysis of treated cells. (C) Schematic representation of the target region of the endogenous PDCD1 gene. One gRNA (blue arrow) was designed to induce a stop codon at residue Q133 (PDCD1_TS1; target C is shown in red). The corresponding protein sequence is shown in gray. (D) Treatment of K562 cells with Pin-point and PDCD1_TS1 gRNA resulted in robust editing at the target site, installing a stop codon at position 133 with about 40% of efficiency. Modified from: (Collantes et al., 2021).
Collantes, J.C., Tan, V.M., Xu, H., Ruiz-Uriguen, M., Alasadi, A., Guo, J., Tao, H., Su, C., Tyc, K.M., Selmi, T., et al. (2021). Development and Characterization of a Modular CRISPR and RNA Aptamer Mediated Base Editing System. CRISPR J 4, 58-68.
2. Development of experimental drugs that modify mitochondrial function for treatment of cancer, obesity, type 2 diabetes, non-alcoholic steatohepatitis (NASH).
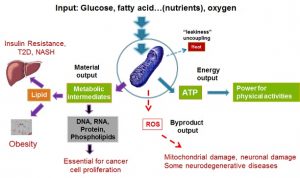
Mitochondria are at the center of the most important medical challenges of our time: obesity, type 2 diabetes, NASH, cancer, and neurodegenerative diseases (see inserted figure). Mitochondria not only provide the majority of energy (ATP) for cellular activity but also are central in producing metabolic intermediates for macromolecule biosynthesis supporting cell proliferation. Moreover, the byproducts of mitochondrial oxidation, reactive oxygen species (ROS), are the main intrinsic causal factor of aging and aging related diseases including neurodegenerative diseases. My laboratory is interested in developing experimental therapeutics that modify mitochondrial activity and function for the treatment of obesity, type 2 diabetes, cancer, and neurodegenerative diseases.
Metabolic diseases: One focus is the development of safe mitochondrial uncouplers as investigational drug leads. Mitochondrial uncoupling is a process by which protons enter the mitochondrial matrix in a way that bypasses the ATP synthase. As a result, mitochondrial uncoupling leads to an increase in lipid oxidation and a reduction in lipid synthesis, an excellent therapeutic strategy for obesity and type 2 diabetes. Our recent discovery of using a safe mitochondrial uncoupler, which is a modified formulation of an FDA-approved drug, for treating type 2 diabetes (Nature Medicine 20, 1263-1269 (2014)) provided proof-of-principle for this approach. Current projects in this direction include developing new mitochondrial uncouplers for other metabolic diseases including obesity, NASH, and alcoholic fatty liver disease. We collaborate with the laboratory of David Augeri, the lead inventor of the Bristol-Myers Squibb type 2 diabetes drug saxagliptin, on the drug discovery program.
Cancer: Almost all cancer cells exhibit aerobic glycolysis (the Warburg effect), which prevents the complete oxidation of glucose or glutamine in mitochondria. As a result, glucose is shunt to pathways for producing metabolic intermediates for biosynthesis of RNA, DNA, and proteins, which are essential for cancer cell proliferation. Mitochondrial uncoupling greatly increases glucose oxidation in cancer cells and deplete the glucose metabolites essential for macromolecule synthesis. This effectively inhibits cancer cell proliferation and represents a novel anti-cancer therapeutic strategy (Alasadi, A. et al, Oncogene 2021, 40(12):2285-2295. da Silva-Diz V, Blood, April 2021; Alasadi, A. Cell Death Dis, 2018, 9(2):215). We are developing new chemical leads for treating a variety of cancers including colon cancer, pancreatic cancer, and leukemia.
Aging and neurodegeneration: Mitochondrial ROS production is very sensitive to the mitochondrial membrane potential (MMP). A slight increase in MMP dramatically increases ROS production while the slight reduction in MMP would reduce the resistance of the mitochondrial electron transport chain, thereby reduces ROS production. Mild mitochondrial uncoupling could be a potential strategy for treating neurodegenerative diseases caused by aberrant mitochondrial ROS production. We are exploring this strategy of intervention.
Mitophagy: Finally, my laboratory has extensive experience and expertise in the study of mitophagy. Mitophagy is the physiological process of removing damaged mitochondria through targeted autophagic degradation. Mitophagy is critical for the maintenance of a healthy population of mitochondria in cells. It is expected that enhancement in mitophagy would promote mitochondrial health and might be the ultimate solution to many diseases of mitochondria origin. My laboratory is engaging basic research in this area, using a powerful system identified in my laboratory, the adipocyte differentiation system. White adipocytes have a unique cellular structure, in which over 99% of the cell volume is occupied by a gigantic lipid droplet, while the rest of the cell structure, including the nucleus, occupies little space. The mature white adipocytes are developed from pre-adipocytes, which are fibroblast-like and contain abundant cytoplasmic components such as mitochondria. Drastic cytoplasmic reorganization and cytoplasm elimination occur during adipocyte differentiation. Tremendous mitochondrial biogenesis occurs in the early phase of adipogenesis, which is required for lipogenesis. We found that massive induction of mitophagy is triggered in later stages of adipogenesis, which is responsible for eliminating mitochondria and other cytosolic components, making room for lipid droplet (PNAS, 106 (47): 19860-5, 2009, and Autophagy, 5(8):1118-30, 2009). This well-characterized model system provides an excellent opportunity to identify the molecular components in mitophagy and the regulatory pathways.