Exploring the diverse biology of prokaryotic immune systems
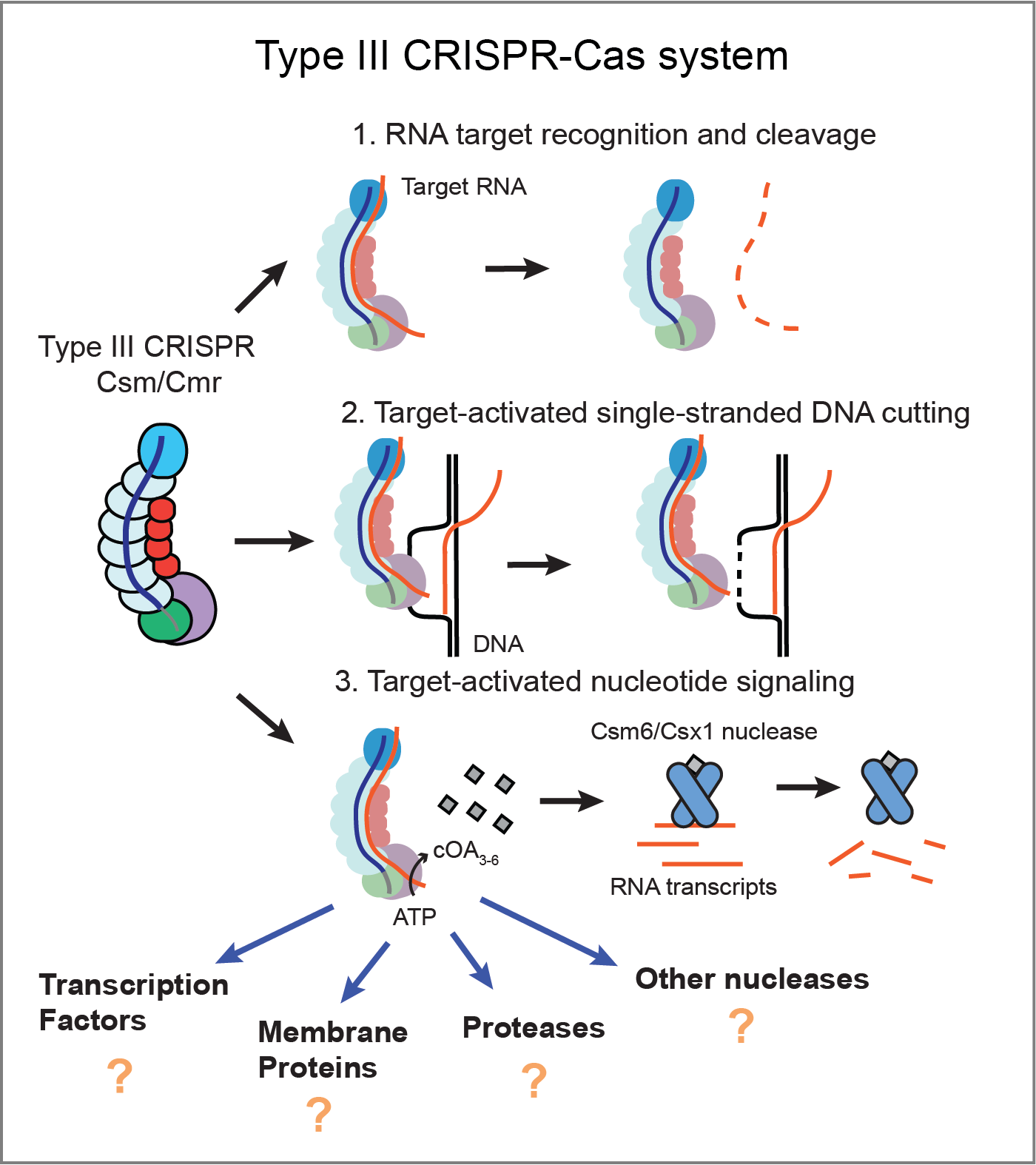
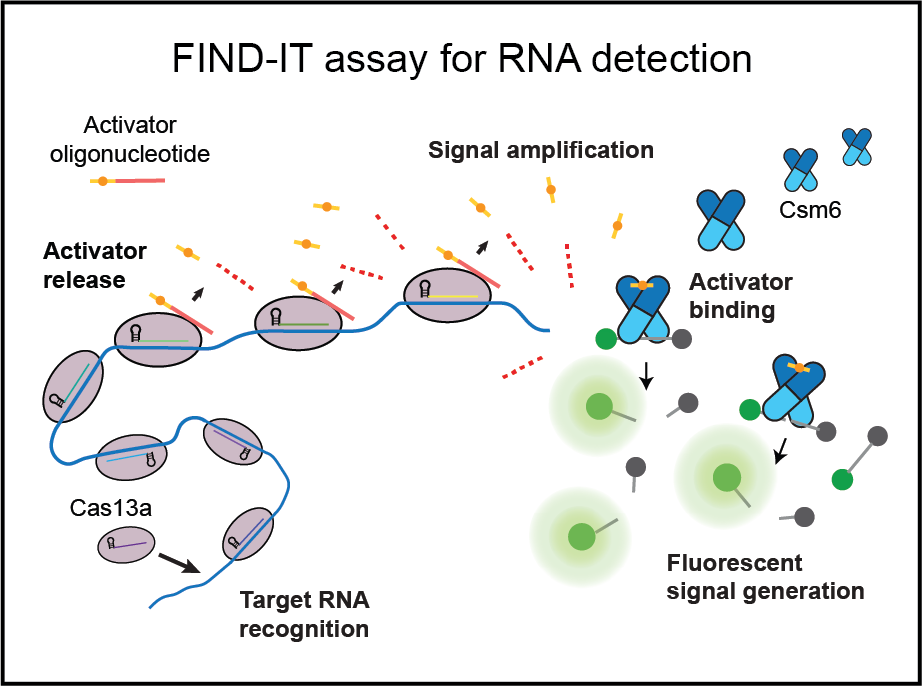
CRISPR-Cas systems are perhaps best known for their applications in genome engineering, in which programmable Class 2 CRISPR effectors, like Type II Cas9 and Type V Cas12, precisely edit DNA in a variety of cell types and organisms. However, Class 1 CRISPR systems, which include Types I, III, and IV, are far more abundant and widespread in nature. During my postdoc, I defined the nucleic acid targeting preference of a Class 1 CRISPR complex called Type III CRISPR Csm. Csm can degrade RNA and DNA, as well as produce signaling molecules that have the potential to activate many different downstream effector proteins, including nucleases, proteases, membrane proteins, and transcription factors. How these work and their role in immunity is not well understood. We aim to explore the diverse functions, mechanisms, and components of the Type III CRISPR signaling pathway. This could reveal entirely new layers of biology, redefining CRISPR as being much more than a pair of molecular scissors. It would also lay the foundation for developing novel tools for cellular engineering.