Research
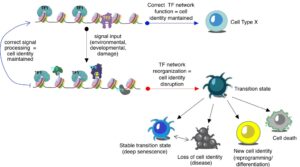
The maintenance of a differentiated cell type (i.e., cell identity) is critical for tissue function, a feature established by cell type-specific transcriptional programs. Yet, the differentiated cell remains a plastic entity, with the ability to functionally reprogram itself in response to incoming signals while simultaneously maintaining its identity. To achieve this, the cell relies on complex TF networks that access, interpret and implement genomic information. TF networks are hierarchically organized, with pioneer TFs playing critical roles in defining cell type- and stimuli-specific enhancer landscapes. Not surprisingly, when TF network function is corrupt, cell identity is destabilized, leading to disease. Our understanding of TF network function and its deregulation during disease development, however, remains poorly understood. Key unresolved questions that will be addressed by my research group are: 1. How do TF networks memorize developmental and environmental signals? 2. How do TF networks maintain epigenome organization across the cell cycle? 3. How does disruption of TF network function promotes disease? The research in my lab aims to answer these questions at the systems level through the combination of high-throughput sequencing approaches, targeted genome editing and computational analyses. Our goal is to use this information to develop better cellular reprogramming strategies for tissue engineering and restoration of cell function as well as the development of epigenetic signatures for diagnostic and prognostic applications.