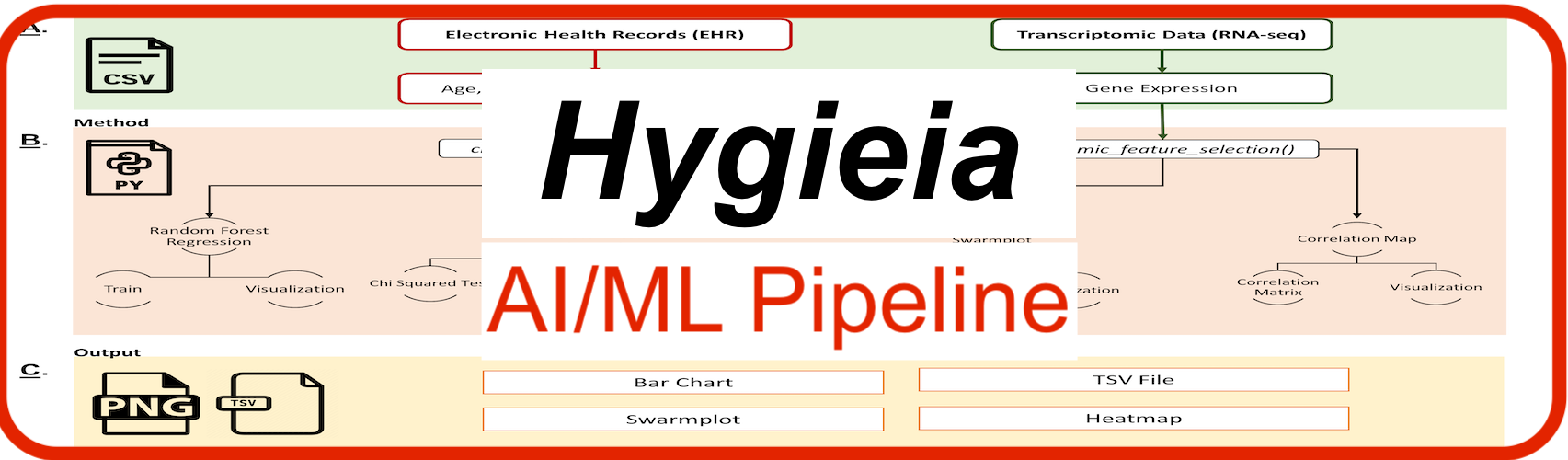
Hygieia: AI/ML pipeline integrating healthcare and genomics data to investigate genes associated with targeted disorders and predict disease.
Due to the advancements in sequencing technologies, genomics data is developing at an unmatched pace and levels to foster translational research. Over ten million genomics datasets have been produced and publicly shared in the year 2022. Genome-wide association studies (GWAS) have remarkably assisted in understanding the genetic basis of human disease by uncovering millions of loci associated with various complex phenotypes. However, GWAS are unable to predict disease and detect all the heritability explained by single nucleotide polymorphisms (SNPs) and can only target specific variants. The rightful use of the artificial intelligence (AI) and machine learning (ML) techniques can accelerate our ability to leverage and extend the information contained within the original data, and model patient-specific genomics data against publicly available annotation repositories for understanding how coding and non-coding genomic variations are connected to disease mechanisms. The grand challenge here is assimilation of genetics into precision medicine that translates across different ancestries, diverse diseases, and other distinct populations with the implementation of effective AI/ML methods. We present first AI/ML ready pipeline i.e., Hygieia., integrating genomics and clinical data to investigate genes associated with the targeted disorders and predict disease with high accuracy. Hygieia can utilize broad dataset sizes with heterogeneous levels of granularity and offer a supervised approach to analyze integrated gene expression and multivariate clinical data. It includes the Random Forest based model for regression analysis and predict without hyperparameter tuning. We trained and tested our model across variable disorders and using diverse datasets. Hygieia is an open-source and simple to use pipeline, which does not strong require computational background to execute.
Publication:
- Degroat, W., Venkat, V., Pierre-Louis, W., Abdelhalim, H., & Ahmed, Z*. (2023). Hygieia: AI/ML pipeline integrating healthcare and genomics data to investigate genes associated with targeted disorders and predict disease. Software Impacts. 100493. (Elsevier).